
Droplet swimmers
In nature, we find large ensembles of active "particles" on many scales - from flocks of animals to microscale systems like plankton and bacteria.
In order to study universal features of such ensembles, we need well controlled model systems lacking the complexity of biosystems, yet showing essential features of self propelled motion.
A very promising system has been observed during the solubilisation of a thermotropic liquid crystal drop in an aqueous surfactant solution (Peddireddy et al., Langmuir 28 (2012) 12426).
A drop expels self propelled micron sized droplets which remain stable over several hours, move freely in 3D, can be mass produced in a simple microfluidics setup and are comparable in size to many classes of single-cell biological swimmers.
Mechanism of the motion
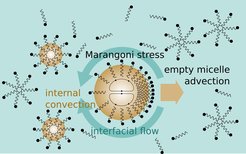
The droplets are pulled forward by Marangoni stresses in the interface, with forces proportional to gradients in surface energy. Outside the lab, these are the forces that make wine and spirits crawl up the walls of a glass or repel a paper boat floating in a water bowl if we drop soap onto the surface. For a droplet in motion, the flow field will advect fresh, empty micelles at the front, while filled micelles are left behind. Since the surfactant coverage of the interface is related to the availability of empty micelles, there will always be more surfactant at the droplet apex. To restore an even coverage, the interface has to relax towards the back, propelling the droplet even further forward. A more rigorous treatment (Herminghaus et al., Soft Matter 10 (2014) 7008) predicts a threshold surfactant concentration beyond which even small positional fluctuations cause self sustained motion. For higher concentrations, the droplet speed increases until it saturates.
Helical swimming
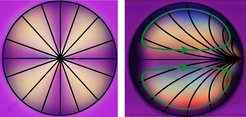
A surprising feature of our swimming droplets is that they swim in a curling/ meandering/helical manner when the constituent liquid crystal is nematic. We can test this simply by observing the swimmers just below and above the nematic to isotropic phase transition at 35°C. This instability is caused by the topological order inside the droplet: liquid crystal molecules are anchored perpendicular to the interface which causes a central "hedgehog" defect at the centre of a moving droplet. If the droplet is in motion, the defect is pulled towards the droplet apex by internal convection.

However, the apex is not a stable position, since the flow field at the interface increases towards the equator. The defect thus is oriented at fixed angle to the droplet axis, which is determined by balancing the forces of the flow field at the interface and the elastic deformation of the liquid crystalline order. The resulting torque adds a constant orbital velocity causing circular trajectories. Superimposed on this is autochemotactic trail avoidance, which causes curling in 2D and helices in 3D (Krüger et al., Phys. Rev. Lett. 117 (2016) 048003).
Chemotaxis and complex geometries
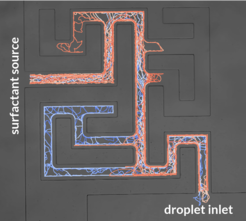
Since the propulsion efficiency depends on the availability of empty micelles, we can expect the droplets to follow surfactant gradients. A demonstration of this feature is their ability to solve microfluidic mazes. In our experiment, we let solid surfactant diffuse into a maze from the exit. Since the concentration will be highest along the shortest path through the maze, droplets will follow this path as soon as the gradient has spread suffciently.
Micellar gradients are also caused by swimmer trails, since available micelles are filled in the wake of a droplet. The limiting time scale for this effect is given by micellar diffusion. For shorter times, droplets exhibit negative autochemotaxis, i.e. they avoid each other's and their own trajectories. We have demonstrated this in a branching microfluidic channel setup. Droplets following each other closely choose alternating branches, an anticorrelation that decays exponentially with time.
Additionally, our droplets swimmers exhibit wall attraction and move in a persistent, non-diffusive manner on the time scale of observation. If they encounter a round pillar, they will be attracted to the pillar wall, but resist the enforced curvature. In an experiment with variable pillar sizes, we were able to observe three regimes: pillars with small diameters, where the enforced curvature is too high and swimmers just bounce off, pillars with very large diameters, where wall attraction dominates everything, and an intermediate range, where wall attraction just outcompetes curvature until the droplet encounters its own trajectory after exactly one pillar orbit and leaves.
3D tracking of buoyant swimmers
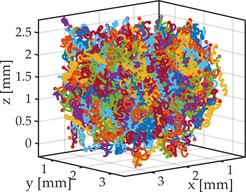
Another serendipitous feature of the 5CB liquid crystal is its density, which is 1.02, barely denser than water. While this is sufficient to have the droplets sediment, we can tune the density of the surfactant solution by partially substituting water with heavy water (D2O) without changing the chemistry of the system, thus going seamlessly from sedimentation to buoyancy to floating. In a custom built light sheet setup, we are able to observe single helical swimmers as well as larger ensembles without the influence of gravity or wall interactions.
Collective behaviour and dimensional confinement
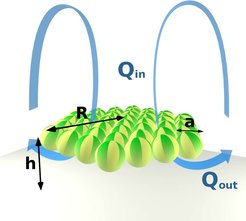
Droplets can collectively sense their state of confinement, purely by hydrodynamic effects. We demonstrate this in an experiment observing large ensembles of droplets in cells varying from very shallow to truly three-dimensional. If the cell barely exceeds the height of the droplets, they only transiently interact with each other, e.g. inhibiting each other's progress in a head-on collision until one of them changes direction. If the cell ceiling is raised, droplets start moving in lines perpendicular to their direction of motion. If allowed to sediment in a large reservoir, droplets arrange in hexagonal clusters, pointing upwards. Since every droplet acts as a pump, this causes a toroidal convection roll enveloping the cluster and lifting it off the cell bottom. The dynamics and morphology of the clusters are determined by the interaction of these large scale flow cells.
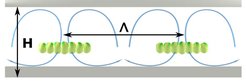
The cluster spacing is set by fitting two convection rolls next to each other
Techniques
Droplets are observed mostly via standard 2D video microscopy. Further information can be extracted by using polarisation filters to probe nematic ordering and adding small colloids to map flow fields via PIV. 3D dynamics is extracted from light sheet scans of fluorescently labelled swimmers. As computational image analysis tools we mostly use customised Python/OpenCV and Matlab interfaces. Microfluidic setups are fabricated in house from externally printed masks.






