Emergent Dynamics in Living Systems
How do complex dynamics and patterns in living systems emerge from stochastic molecular interactions in the cell, how are they coordinated at the population/tissue level, and what role do environmental constraints and interactions play in shaping and maintaining them?
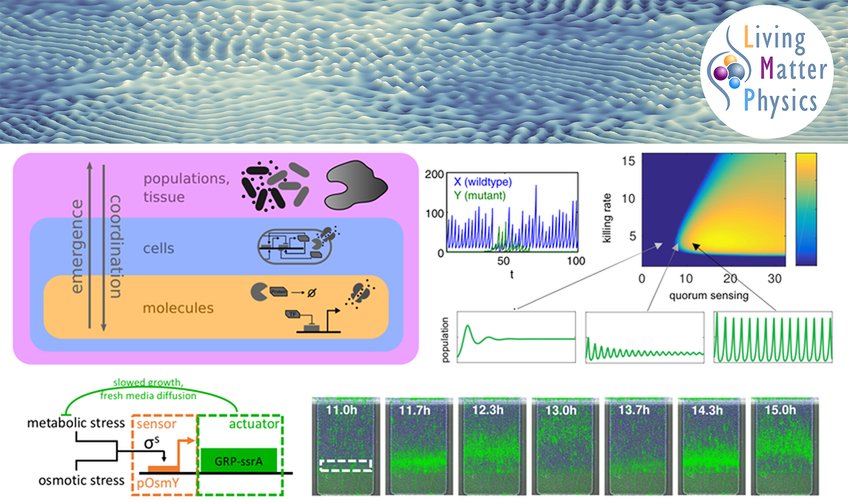
Complex dynamics and pattern formation which arise from the interaction of different components or agents are not only ubiquituous in nature but also essential to many biological functions in living systems that range from bacterial populations to mammalian tissues. In collaboration with quantitative experimentalists, our research at the interface of physics and biology combines concepts from nonlinear dynamics and active matter with mathematical modeling to disentangle these processes. We are particularly interested in the collective effects of proliferation, i.e. cell growth and division, a key process without which life would not be possible, but also other cellular activities such as motility and metabolism. Our goal is to extract the fundamental mechanisms that are responsible for observed behavior and at the same time provide strategies for controlling biological systems for biotechnological and medical purposes.
We investigate models of cellular aggregates that undergo self-organization due to an entanglement of physical processes such as mechanics and chemical transport with the single-cell dynamics such as cell division, gene regulation and metabolism. Using descriptions of varying degrees of detail (stochastic, determinstic, agent-based, continuous), we consider cell-to-cell interactions and internal regulation leading to spatial organization, patterning, competition between multiple strains and evolution. The results of these investigations are broadly applicable to bacterial populations (biofilms) and tissues, with possible implications for, e.g., antibiotic resistance, synthetic biology (engineered microbial systems), embryogenesis, tissue formation and maintenance, and tumor dynamics.
Mechanical interactions due to growth and division
Current members
Lukas Hupe (Postdoc)
Jonas Isensee (Postdoc)
Ilias-Marios Sarris (Ph.D. student)
Torben Sunkel (Ph.D. student)
Tobias Büchner (M.Sc. student)
Jonas Willms (M.Sc. student)
Finn Albrecht (M.Sc. student)
Lucas Reum (M.Sc. student)
Philipp Rademacher (B.Sc. student)
Associated
Yoav Pollack (former Postdoc of LMP department)
Patrick Zimmer (joint M.Sc. student with Yoav Pollack)
Samantha Lish (former OxCam PhD student)
Alumni
Dorian Marx (Bachelor student, joint with J. Agudo-Canalejo)
Florian Aubermann (intern, Heidelberg University)
Thomas Boukéké-Lesplulier (intern, ENS Lyon)
Christina Goss (intern, Northwestern University)
Lisa Graßmel (B.Sc. student)
Carl Hormes (B.Sc. student)
Leif Peters (M.Sc. student)