Pattern Formation in Biological Systems
A) Pattern formation in Dictyostelium discoideum
I am interested in the experimental and theoretical aspects of pattern formation in colonies of Dictysotelium discoideum (D.d.) cells. The slim mold D.d. is a valuable model organism to study pattern formation and development in biology. When deprived of nutrients, D.d. cells aggregate by a chemotactic response to the chemoattractant cAMP and then develop to form multicellular fruiting bodies. The cells initiate the process by the spontaneous release of cAMP pulses. The other cells respond to the attractant stimulation by moving towards its source and by relaying the signal. As a result, circular and spiral wave patterns form with time, which have a periodicity of few minutes. The main focuses of our investigations are the following:
A-1) Pattern formation in the presence of an external flow
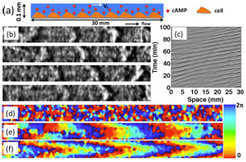

In the natural environment, populations of starving D.d. may experience fluid flows that will profoundly alter the wave generation processes as we show here. Although in nature these wave processes occur in 3D, a first understanding may be gained in a quasi 1D geometry. We investigate spatial-temporal dynamics of a uniformly distributed population of D.d. cells in a flow-through narrow microfluidic channel. As shown in the Figure 1, the starved amoebae are attached to the surface of the channel and exposed to an external fluid flow that advects cAMP molecules downstream. This transport anisotropy in the extracellular medium induced macroscopic wave trains that differ profoundly from conventional excitation waves in this system. In our theoretical investigations, we focus on the convectively unstable regime, where a perturbation applied to the system dies out in the laboratory reference frame, while it grows in a moving one. By fixing the upstream boundary condition the system becomes unstable, producing a trigger wave that travels upstream, and a wave train propagating downstream. The trigger wave is absorbed when it reaches the upstream boundary, then the system destabilizes again, and the phenomenon repeats. In our 2-D simulations the trigger wave propagating against the flow has a triangular shape, similar to concentration profiles exhibiting a cusp in auto-catalytic advection reactions.
A-2) Pattern formation in the presence of external obstacles
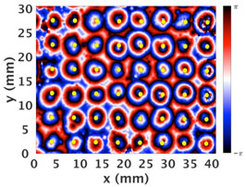
In their natural habitat, populations of starving cells are exposed to spatial heterogeneities that will profoundly influence the processes of wave generation and propagation. In nature the obstacles are randomly distributed in 3D, but as a first step towards understanding, we look at a simpler system of cells in a 2D geometry with a periodic arrangement of obstacles. Our quasi-2D geometry consists of a regular array of millimeter-sized pillars that control spatio-temporal dynamics of a population of uniformly distributed D.d. cells. As shown below, these spatial heterogeneities induce circular waves centered on the posts that trigger chemotactic cell movement towards the pillars. This leads to the formation of periodic Voronoi domains that reflect the periodicity of the underlying macro-pillar array.

B) Synthetic biology
In this project, we aim to attach beating isolated cilia/flagella to a substrate and build a cilia carpet. A challenge is to establish a protocol in which the flagellum binds only from one end to the substrate. Up to now, we have successfully isolated and reactivated flagella from the green algae Chlamydomonas reinhardtii. Flagella are slender cellular appendages whose regular beating propel cells and microorganisms through aqueous media. The flagella from Chlamydomonas are approximately 12 µm long and 0.2 µm thick. The beat is an oscillating pattern of propagating bends generated by dynein molecular motors. An example of the beating pattern of an isolated flagellum and the corresponding curvature waves are shown in the figure below. The white and blue star-like trajectories depict the position of the basal and distal ends of the axoneme.



